AffyExtensions - An R package
Written by Ben Bolstad
email bmb@bmbolstad.com
|
|
Special Note For the vast majority of people you should look at affyPLM for the functionality you previously found in AffyExtensions
|
What?
The package AffyExtensions contains a set of routines that can be called from within R. It extends the functionality of the R-packages: affy and affyPLM. These are themselves components of the Bioconductor project.
|
Why?
You might ask why I don't just seek to get these functions included into the base affy package. The reason is that one of the main goals of that package is to be general and flexible, allowing one to add new methods using the framework of the package. The code in AffyExtensions will in general be less flexible that that in the main package, but I trade that off for speed of execution. Some code that makes it into this package will eventually be integrated into the main packages. A fast RMA based upon the code in this package was integrated into the main affy package as of release bioconductor 1.1.
Another purpose of this code is as a testing ground for stability purposes. Because of this things may occasionally change drastically, eg 0.3 and 0.4 series are completely different. In otherwords things change rapidly in AffyExtensions with much speculative work.
|
How does AffyExtensions relate to Bioconductor, affy and affyPLM?
For the most part AffyExtensions has been Bioconductor compliant. Meaning that it would work satisfactorily and in combination with Bioconductor packages. AffyExtensions has also served as a place for some experimental and sometimes highly speculative work. Some routines have eventually become stable or well developed enough that they have transitioned into the base affy package: examples include RMA.C() of AffyExtensions 0.3.X which became the rma() routine in affy at the 1.1 release, also parsing routines developed in the 0.5 series of AffyExtensions have become the default routines in the affy 1.3.X series.
affyPLM is a BioConductor package that started with a major portion of AffyExtensions 0.5-14. It is a direct port of that version of the package to R-1.8 with a few routines removed. This package concentrates on Probe Level Modelling procedures. Later on further developments from AffyExtensions were integrated into affyPLM. At the conclusion of the AffyExtensions 0.7-X series most of the major functionality had been integrated into affyPLM and subsequent development was carreid out on affyPLM rather than AffyExtensions.
To perhaps best understand the history and roadmap of the packages this diagram clarifies some of the interdependencies. Please click for a larger version
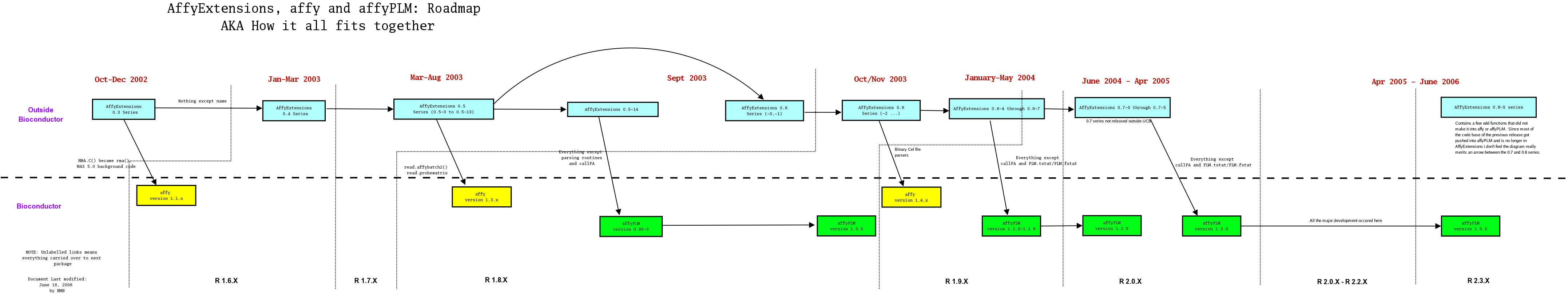
|
Short history of changes.
| Approximate Dates | Version | Short Description |
| Jun - Sep, 2002 | pre 0.3 | A collection of loose C and R code |
| Oct - Dec, 2002 | 0.3 Series | A package supplying RMA.C(), which later became rma() in the affy package. | Jan - Mar 2003 | 0.4 Series | Three Step methods. Robust linear modeling at the probe level. |
| Late Mar 2003 - Sep, 2003 | 0.5 Series | New backgrounds, dependence on R-1.7.0 and later, more general probe level modeling |
| Sep 2003-May 2004 | 0.6 Series | Modified PLMset object, many additional features |
| Jun 2004-Apr 2005 | 0.7 Series | Never released outside UCB. Lots of work particulary in how the model fitting algorithm was implemented and the range of possible models that could be fit. Became the base for affyPLM at 1.3.X |
| Jun 2006 - Present | 0.8 Series | Implement the few things that never quite made it into affy or affyPLM. Large dataset support |
|
How do I get it? and what do I need?
For most people affyPLM will provide all the functionality they are expecting and if you have that there is no need to install AffyExtensions.
| Version 0.8 series works with BioC 1.8. You can download this from www.bioconductor.org. |
| Linux/Unix and the like | Windows | Date | Comments |
| AffyExtensions_0.8-0.tar.gz | Not currently available | June 16, 2006 | Restarting the package with a few things that did not make it into other packages. Compatible with BioC 1.8 and R 2.3.X |
| AffyExtensions_0.8-1.tar.gz | Not currently available | Aug 10, 2006 | Extremely large dataset support for computing RMA expression values. Built on top of the BufferedMatrix package. |
| AffyExtensions_0.8-2.tar.gz | Not currently available | Jun 19, 2007 | Update justRMALite() to compatibility with BioC 2.0 |
If you want access to the large dataset functionality you will need the BufferedMatrix package. Currently you must download and build the sources yourself from the BioC svn archive. BioConductor svn repository (user: readonly, pass: readonly)
|
What is this "large data support" all about?
As of AffyExtensions 0.8-1 a function justRMALite has been introduced that can process extremely large numbers of CEL files to produce RMA expression values. While justRMA will also process large numbers of CEL files it is also extremely memory hungry. justRMALite uses temporary files to store interim data and calculations so it has a lower memory overhead. The following plot illustrates the running time differences between the functions:
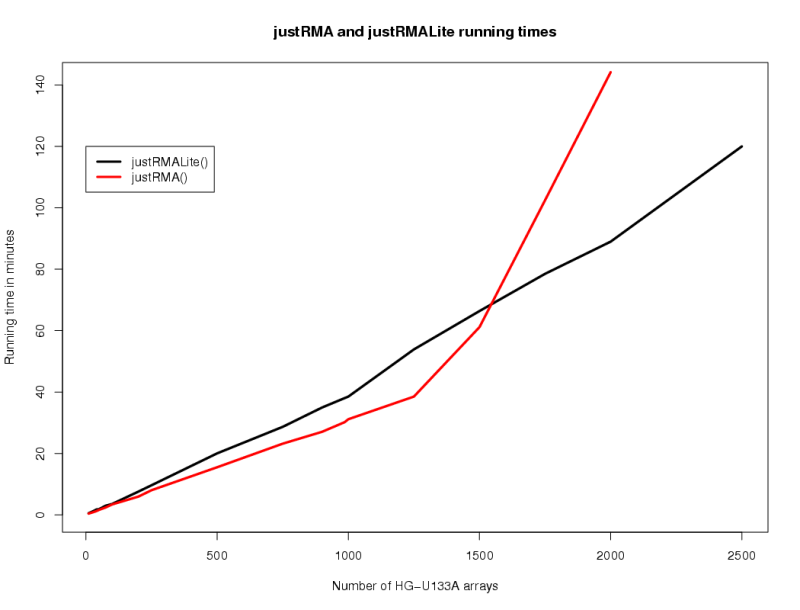 This benchmark was established based on a machine with the following hardware/software configuration:
This benchmark was established based on a machine with the following hardware/software configuration:
| Component | Description |
| Processor | AMD X64 3800+ |
| RAM | 3 GB |
| SWAP | 6 GB |
| Operating System | Linux (Fedora Core 5 with 2.6.17-1.2157_FC5 kernel) |
| Free disk space | A lot !!! |
| R | 2.3.1 |
| AffyExtensions | 0.8-1 |
| affyPLM | 1.9.11 |
| affy | 1.11.4 |
| affyio | 1.1.7 |
| BufferedMatrix | 0.1.12 |
| Biobase | 1.10.0 |
All the U133A CEL files used were text format (sometimes called version 3) although both functions will also handle the binary format. Because the test machine has a large amount of RAM we see better performance from justRMA up to about 1500 CEL files at which point justRMALite becomes faster. Based on the shape of the curve for justRMA we can estimate the point at which the program starts seriously dealing with swap memory rather than RAM (approximately 1250). On machines with less RAM the point at which justRMALite becomes more speed efficient will be lower.
|
What documentation is there?
Documentation for the 0.4 and later series are in the vignettes. Type openVignette() to read vignettes.
|
Any other questions?
Send me an email at bmb@bmbolstad.com
|
|
|

 This benchmark was established based on a machine with the following hardware/software configuration:
This benchmark was established based on a machine with the following hardware/software configuration: